Our research in
Foldamer chemistry
SYNTHESIS OF FOLDED AND SELF-ASSEMBLED ARCHITECTURES MIMICKING PROTEINS
One of the main line of research in the Guichard group is the construction of new synthetic folded systems mimicking biopolymers (‘foldamers’) coupled to the exploration of the interplay between folding and molecular recognition properties. Using proteins as inspiration, our group has pioneered the development of peptidomimetic foldamers without amino acids by showing that aliphatic N,N’-linked urea oligomers form well defined and remarkably stable helical secondary structures reminiscent of the α-helix. Our current projects focus on the development of more sophisticated and functional architectures including peptide/oligourea hybrids, the formation of new folding patterns, the design of composite proteins (tertiary structures), and of nanostructures formed by controlled foldamer assemblies (quaternary structures).
Interfacing peptide and oligourea foldamers
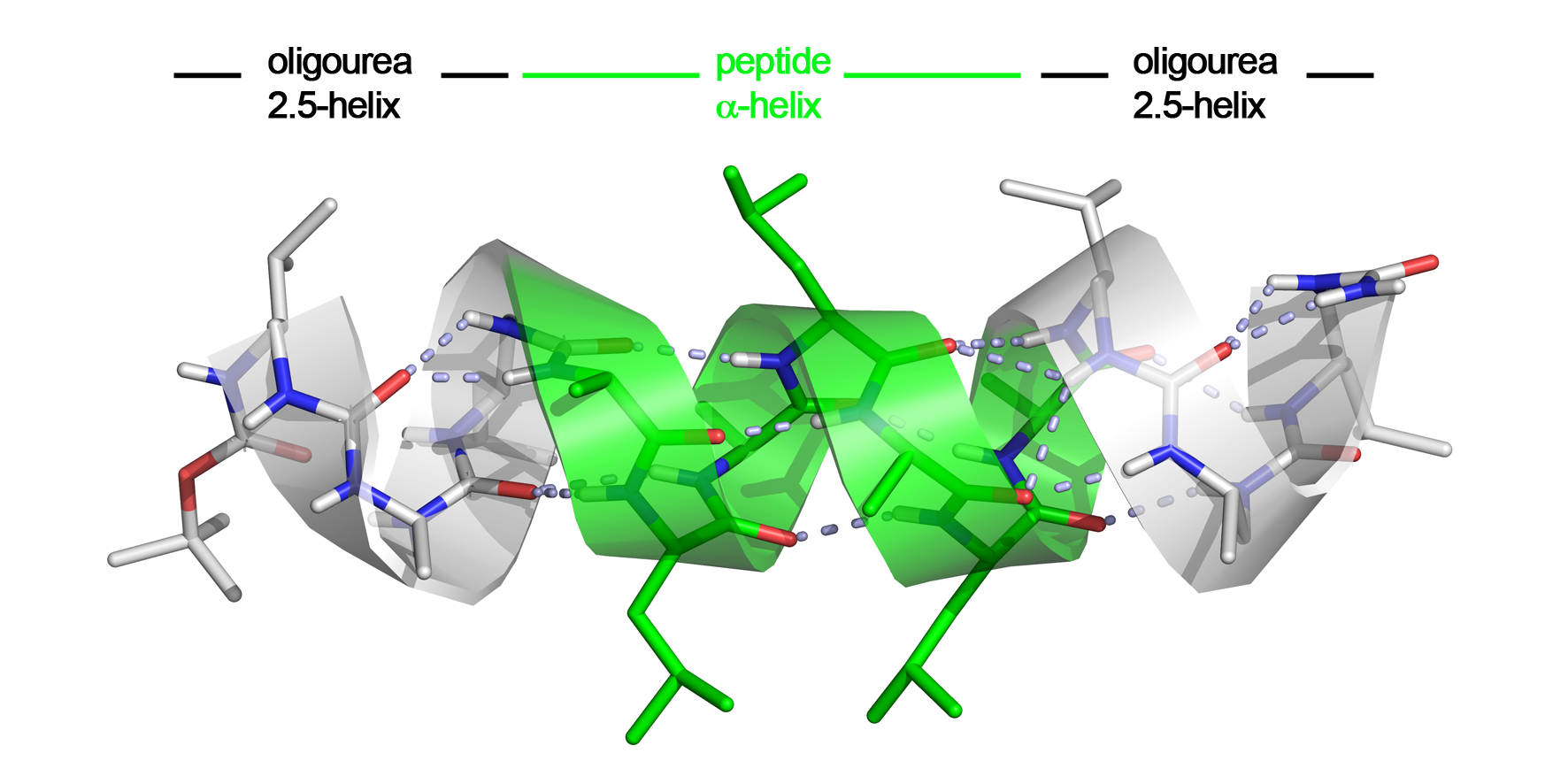
We recently discovered that peptide and oligourea helices can communicate when embeded in a single strand to generate unique helical hybrid structures, suggesting possible applications for mimicking bioactive α-helical peptides (ACIE 2015a, C.R. Chimie 2016 and one patent application). The rationale for interfacing urea-based foldamers and α-peptides was that the oligourea helix shares a number of features with the α-helix such as helix polarity and pitch. In this project, we have investigated the ability of aliphatic oligoureas fused to short peptide segments to nucleate α-helical structures. We found that the resulting chimeras were fully helical in the solid state as well as in polar solvents, with as few as three urea units sufficient to propagate an α-helical conformation in the fused peptide segment. The remarkable compatibility of α-peptide and oligourea backbones, along with the simplicity of the technology is of particular significance for future applications of foldamers in biology and medicine including the inhibition of protein-protein interactions (PPIs).
Funded by :
Composite proteins containing foldamer segments
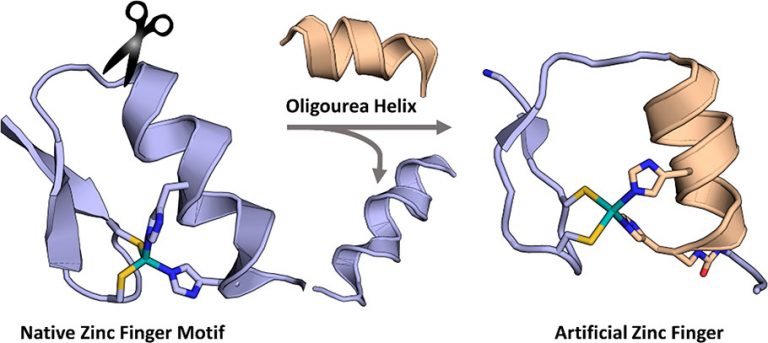
The purpose of this project is to explore the chemical synthesis, folding, and function of composite proteins in which a folded peptide segment (e.g. an α-helix) is replaced by an unnatural counterpart (e.g. a foldamer). The similarities in screw sense, pitch, and polarity between peptide α-helices and oligourea 2.5-helices suggested to us that a tertiary structure could be retained when swapping the two backbones in a protein sequence. We have started this project by selecting the Cys2His2 zinc finger motif as a model protein fold and replaced the 10-residue long original α-helical segment in the third finger of transcription factor Egr1 by an oligourea sequence bearing two appropriately spaced imidazole side chains for zinc coordination. We showed by spectroscopic techniques that the composite zinc finger retains the zinc coordination properties of the native protein and adopts a folded structure in which the native β-sheet arrangement of the peptide region and global arrangement of DNA-binding side chains are preserved. Titration in the presence of the Egr1 target DNA sequence supports binding to GC bases as reported for the wildtype zinc finger.
Funded by :
Engineering foldamer quaternary structures in aqueous solution
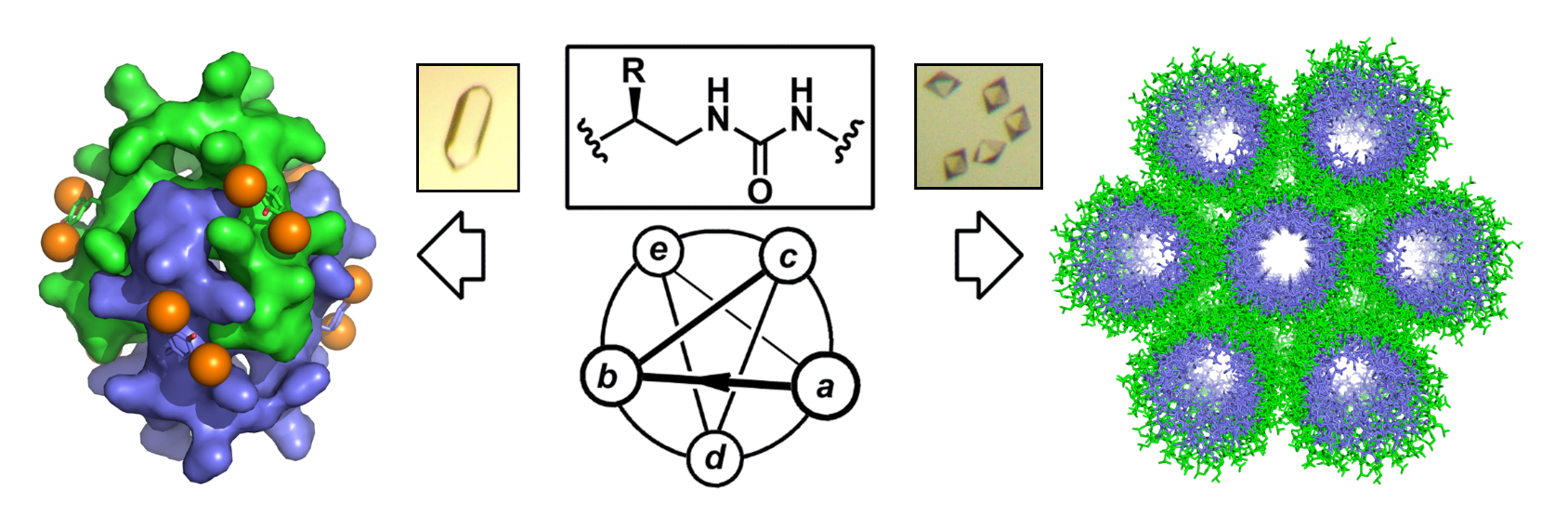
The aim of this project is to create ensembles of complex synthetic and functional nanoscale molecular architectures with new compositions and shapes by using (i) non-natural sequence-specific oligomers with high folding fidelity, and (ii) a fabrication process in aqueous conditions mimicking those at work in proteins (i.e. hierarchical structure formation). The high resolution three-dimensional structures (i.e. extended channel-type assembly and discrete helix bundle) published in Nature Chemistry demonstrated our ability to orient the assembly process towards compact or extended nanostructures in aqueous conditions by sequence manipulation and redistribution of charged side chains at the surface of the helix. In this work, X-ray crystallography served as a major analytical technique (in combination with solution studies) to characterize the resulting assemblies at atomic resolution, and learn about sequence-to-structure relationships as a mean to refine the design of foldamer sequences (See also Chem Commun 2016, Chem Sci 2016, JACS 2016).